Abstract
Introduction. In myelodysplastic syndromes (MDS) TP53 mutations are present in <10% of newly diagnosed patients (pts) and in >30-40% of those with therapy-related MDS. Loss of the TP53 locus represents one of the most powerful risk factors. TP53 mutations frequently result in loss of trans-activating function and stabilization of mutant p53 proteins, leading to their increased accumulation at cellular level (detectable by immunohistochemistry). However, p53 stabilization in cancer can also be observed in the presence of wild-type TP53 alleles, frequently associated with altered expression of p53 regulatory elements (Nat Rev Cancer 2014;14:359-370). In this study we evaluated the prevalence and clinical implications of p53 dysfunction in a large MDS population
Methods. We retrospectively studied a cohort of 998 MDS pts receiving best supportive care (BSC, n= 495), hypomethylating agents (HMAs, n= 198) and allogeneic transplantation (HSCT, n= 305). We used massively parallel sequencing to examine tumor samples somatic mutations in 30 recurrently mutated genes in myeloid neoplasms (JCO 2016;30:3627-3637). To detect p53 protein expression, paraffin sections were stained with DO-1 antibody (Santa Cruz Biotechnology, Santa Cruz, CA),according to a standardized technique (JCO 2011;29:1971-1979). Recursive partitioning was used to generate a hierarchical model for overall survival integrating clinical and genetic characteristics (NEJM 2017;376:536-47)
Results. TP53 mutations were detected in 11% of pts. Pts with TP53 mutations of had a paucity of other gene mutations; TP53 mutations were infrequent in pts with splicing mutations vs pts with wt splicing factor genes (P=.002). TP53 mutations were associated to advanced disease according to R-IPSS (P=.02) and poor/very poor cytogenetic risk (P<.001). Survival of pts with TP53 mutation was shorter than wt (15 vs 66 months, P < .001) and it retained significance in multivariable model. None of the sequentially analyzed samples showed a disappearance of the mutant clone or emergence of new clones, suggesting an early occurrence of TP53 mutations. A reduction in mutant clone correlated with response to HMAs and HSCT, however clones increased in non-responders and persisted at disease relapse.
In order to define pts with p53 dysfunction, a threshold of 2% of positive cells (p53++/dark brown, nuclear staining) was used, based on the correlation with TP53 mutational status (negative and positive predictive value for the presence of mutation were 96% and 79%, respectively) and the capability to capture an high risk of leukemic evolution. The concordance between the reviewers of bone marrow histology resulted in >95% agreement (P<.001) for recognition of percentage of p53++ cells. Focusing on MDS with increased p53 expression (17% of the whole population), subjects carrying TP53 gene mutations showed a survival comparable to that of pts with wild type TP53 gene (15 vs 17 months, P=ns).
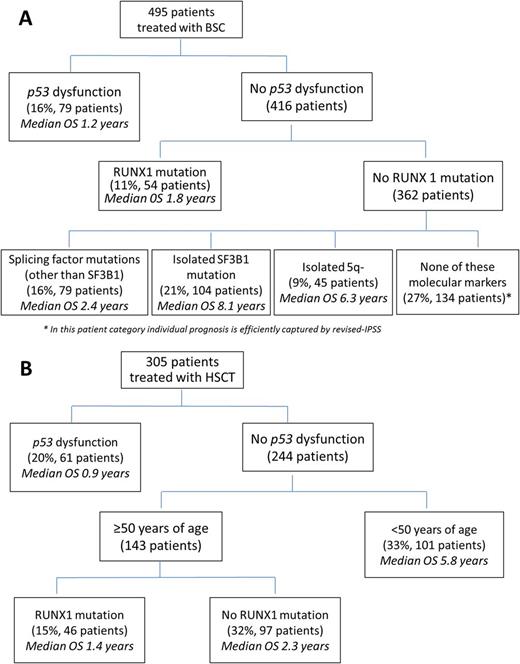
We than defined a disease category, i.e. MDS with p53 dysfunction, including pts with TP53 mutations and/or p53 hyperexpression. We did a recursive partitioning analysis considering in addition hematological features and gene mutations as variables. In pts receiving BSC, p53 dysfunction was the most relevant independent prognostic factor associated with increased risk of disease progression and short OS (p=0.001)(Figure 1A). In pts treated with HMAs, p53 dysfunction was associated with lower complete remission rates (21% vs 25%; p=.038) and higher probability of relapse (p=.01). In MDS who received HSCT, the presence of p53 dysfunction was the most important prognostic variable associated with the risk of transplantation failure due to high risk of disease recurrence (p=0.002)(Figure 1B).
Conclusions. TP53 mutations occur in about 10% of MDS cases, while wild-type p53 dysfunction due to non-mutational p53 abnormalities appear to be rather frequent in these diseases. MDS with p53 dysfunction appear to be a distinct clinical entity characterized by poor prognosis due to high risk of disease evolution and low probability of response to conventional treatments. TP53 genotyping with evaluation of p53 protein expression may contribute to the precise assessment of p53 status in MDS, thus leading to the identification of cases candidate to a p53-based therapy.
Figure 1 OS based on recursive partitioning analysis in A) 495 MDS pts receiving BSC B) 305 pts treated with HSCT
Santini: Janssen: Consultancy, Honoraria; Abbvie: Consultancy, Membership on an entity's Board of Directors or advisory committees; Novartis: Honoraria; Otsuka: Consultancy; Celgene: Honoraria, Research Funding; Amgen: Membership on an entity's Board of Directors or advisory committees. Santoro: Merck: Membership on an entity's Board of Directors or advisory committees; Bristol-Myers Squibb: Membership on an entity's Board of Directors or advisory committees. Rambaldi: Novartis, Roche/Genentech, Amgen, Italfarmaco: Consultancy; Novartis, Amgen, Celgene, Sanofi: Other: Travel, Accomodations, Expenses.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal